Optimal RNA Purity with TRIzol®
TRIzol®-based reagents have long been considered the best method for isolating high-quality, pure RNA from cells or tissues. TRIzol® can be used as both a storage medium and extraction solution for RNA, and it preserves the sRNA quality, integrity, and quantity. TRIzol® can be used to lyse a variety of biological samples including cells, tissues, serum, and microbes, making it an extremely versatile reagent. TRIzol® can be used to recover RNA from very small or very large amounts of input samples and is also an effective ribonuclease (RNAse) inhibitor, preventing degradation of RNA during the purification process.
Using TRIzol® for RNA Isolation
TRIzol® is a low-pH solution of phenol and guanidine isothiocyanate ideally designed for the separation of RNA, DNA, and protein from various biological inputs. At a low pH, all three components will be isolated into separate layers from each other. The guanidinium isothiocyanate, a salt, is a chaotropic agent that denatures proteins in the sample.
After solubilization and homogenization of a sample in TRIzol®, RNA, DNA and protein are differentially extracted in a test tube by the addition of a phase separation reagent such as chloroform, BCP or BAN. An aqueous solution at the top of the tube contains the RNA. An interphase contains mostly DNA as well as partially denatured proteins and lipids. The phenol in TRIzol® is an organic compound in which protein but not DNA or RNA is soluble, and this is the bottom layer in the tube. The phenol in TRIzol® is commonly colored pink, making the three layers easily distinguishable from each other (Figure 1).
Following phase separation, the RNA is collected by alcohol-based precipitation when the aqueous phase is mixed with isopropanol. After several incubation and centrifugation steps, the resulting pellet of purified RNA is washed in an ethanol solution, air-dried, and resuspended in an aqueous buffer such as RNAse-free water or TRIS buffer.
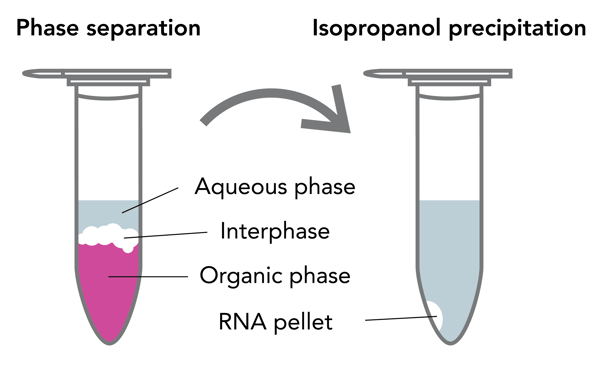
Figure 1. Phase separation using phenol and chloroform, demonstrating the three layers. The aqueous phase contains RNA; the interphase contains DNA, partially degraded proteins, and lipids; and the organic phase contains DNA and denatured proteins. The aqueous phase is transferred to another tube where the RNA is extracted by isopropanol precipitation.
Troubleshooting TRIzol® RNA Purification
Although extraction of RNA with TRIzol® results in high-purity RNA, it comes with some challenges. The many steps involved in TRIzol®-based RNA extraction mean that it is not adaptable to high-throughput experiments performed on automated liquid handling instruments. Other challenges to using TRIzol® include the use of chloroform for phase separation and long centrifugation times to precipitate and purify the RNA.
Characteristics of the input sample must be taken into account when preparing your experiment that will use the TRIzol® method of RNA extraction. When a small number of cells are used, microRNAs low in C-G nucleotides are inefficiently extracted, as are microRNAs with a secondary structure.
Inefficient cell or tissue lysis is also a common problem that must be addressed when using TRIzol®. There must be an ample amount of TRIzol® compared to input sample in order for lysis to efficiently occur. To be sure, researchers can increase the volume of TRIzol® over what is recommended in the protocol. Alternatively, performing mechanical lysis of the sample in TRIzol® prior to purification will yield robust total RNA. Some options for mechanical lysis are bead beating tubes or homogenization (Figure 2). Incorporating both mechanical lysis and increasing the volume of TRIzol® will ensure a higher quality and quantity of RNA is isolated.
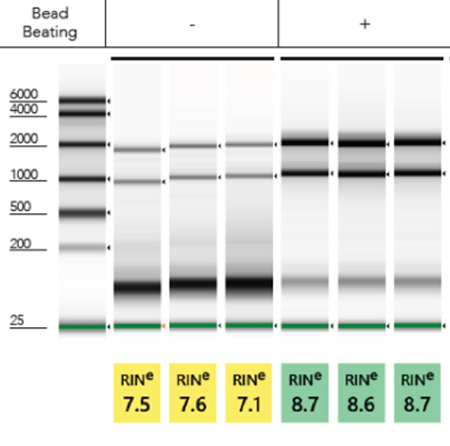
Figure 2. Total RNA isolated from E. coli cells (in triplicate) with (+) or without (-) bead beating mechanical lysis. Bead beating yield robust 28S/18S ribosomal bands and high ribosomal integrity numbers (RINe). Samples analyzed by Agilent Tapestation® 2200.
During the removal of the aqueous phase following phase separation, cross-contamination of the RNA with proteins, lipids, DNA, and phenol can occur (Figure 3). To identify phenol contamination, a characteristic absorbance maxima at 270 nm will be seen when quantifying an RNA sample on a spectrophotometer (Figure 4). Genomic DNA contamination can be detected by unexpectedly high yields on a spectrophotometer and/or the appearance of high molecular weight bands or a smear on an agarose gel (Figure 5).
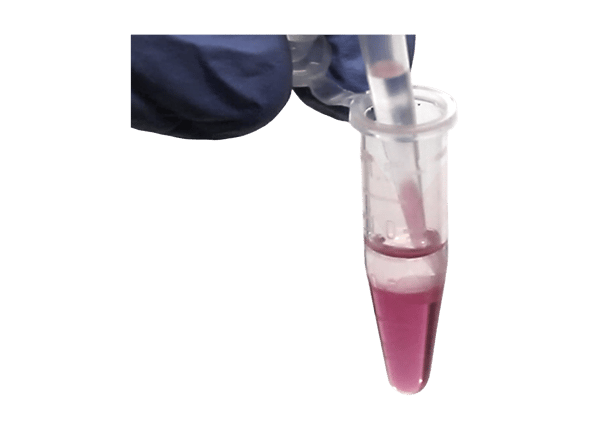 Figure 3. During the transfer of the aqueous phase following phase separation, the organic phase material containing DNA, proteins, lipids, and phenol can contaminate the RNA in the aqueous phase if the aspiration is not performed carefully.
Figure 3. During the transfer of the aqueous phase following phase separation, the organic phase material containing DNA, proteins, lipids, and phenol can contaminate the RNA in the aqueous phase if the aspiration is not performed carefully.
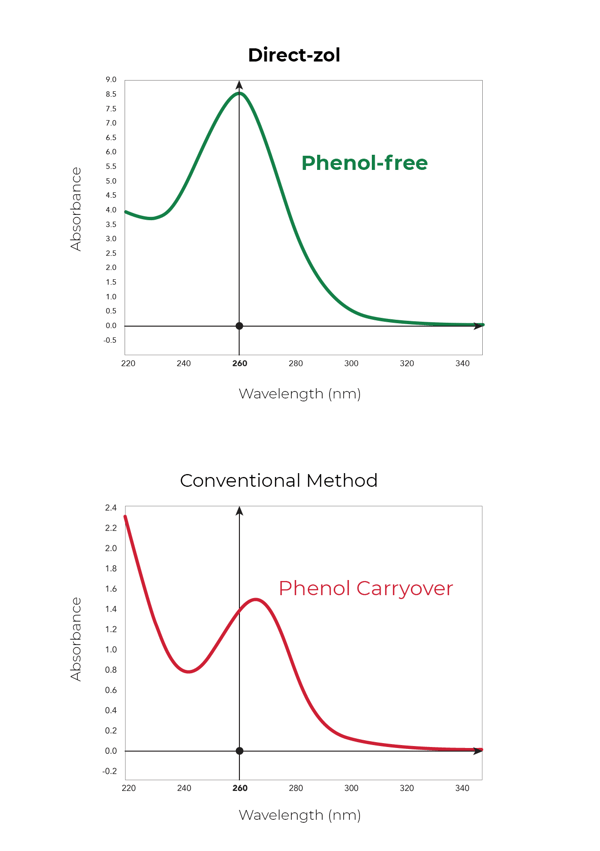 Figure 4. A peak with an absorbance maximum at 270 nm indicates phenol contamination of the aqueous phase. Graphs demonstrate the effect of phenol contamination on absorbance spectra (left panel) versus a non-contaminated, phenol-free sample (right panel).
Figure 4. A peak with an absorbance maximum at 270 nm indicates phenol contamination of the aqueous phase. Graphs demonstrate the effect of phenol contamination on absorbance spectra (left panel) versus a non-contaminated, phenol-free sample (right panel).
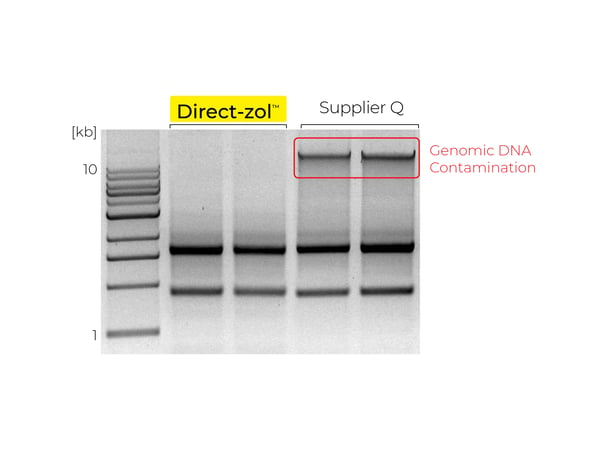 Figure 5. DNA contamination of RNA is seen as a high molecular weight band or smear, indicated by the red box, from TRIzol® samples extracted with a Supplier Q kit. DNA contamination is not observed in RNA extracted from TRIzol® with Zymo Research’s Direct-zolTM RNA kit.
Figure 5. DNA contamination of RNA is seen as a high molecular weight band or smear, indicated by the red box, from TRIzol® samples extracted with a Supplier Q kit. DNA contamination is not observed in RNA extracted from TRIzol® with Zymo Research’s Direct-zolTM RNA kit.
Alternatives to TRIzol® for RNA Extraction
Nowadays, many kits and phenol-free methods for RNA extraction exist, but some labs continue to use TRIzol®. They may have samples frozen in TRIzol® or prefer the reagent due to its high-purity RNA output. However, there is still a need for a method of RNA extraction that is faster and more reliable than a traditional TRIzol® protocol but provides the same high-purity RNA and can be used with a wide range of input sample volumes.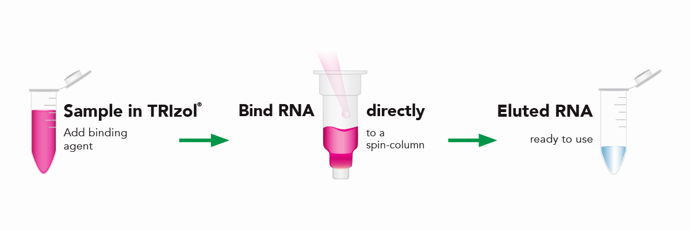 Figure 6. Zymo Research’s Direct-zol™ RNA workflow eliminates the phase separation or isopropanol precipitation steps, resulting in a simple workflow in which the sample in TRIzolTM can be bound directly to a spin column and RNA can be extracted in as little as 7 minutes.
Figure 6. Zymo Research’s Direct-zol™ RNA workflow eliminates the phase separation or isopropanol precipitation steps, resulting in a simple workflow in which the sample in TRIzolTM can be bound directly to a spin column and RNA can be extracted in as little as 7 minutes.
Zymo Research’s Direct-zol™ RNA Kits are designed to purify any sample stored or prepared in TRIzol®. The Direct-zol™ technology eliminates DNA and phenol contamination and does not involve the time-consuming phase separation and precipitation steps from a traditional TRIzol® protocol. Instead, the sample in TRIzol® is added to an RNA-binding spin column, which is then washed and then the RNA is eluted (Figure 6). Direct-zolTM RNA Kits can extract as low as picogram quantities of RNA (single-cell amounts) up to 100 µg RNA per sample (Figure 7). This RNA is ready for use in sensitive downstream applications such as real time PCR/quantitative PCR (qPCR) and next generation sequencing.
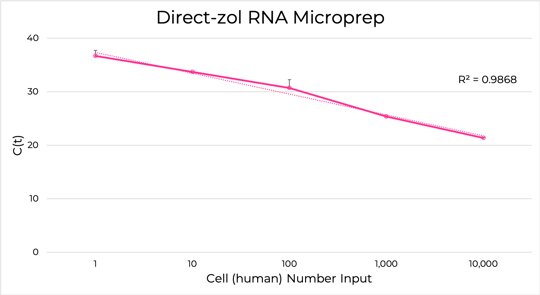
Figure 7. Total RNA purified using the Direct-zol™ RNA Microprep kit for a range of input cell numbers, from 104 cells to a single human epithelial cell, as detected by RT-qPCR for beta-Actin. Each C(t) value represents two averaged samples.
Try Zymo's Direct-zol™ RNA Kits Today!