MUST-KNOW TIPS FOR RNA EXTRACTION FROM TRIZOL®
RNA extraction is among the first steps in studying gene expression using a variety of different methods, including northern blotting, transcriptome analysis, and reverse transcription-PCR. These techniques have become ubiquitous across myriad biomedical fields, from cancer to heart disease to neurodegeneration, positioning RNA purification as a foundational technique for these rapidly growing, highly-active research areas.1-3
HOW RNA PURIFICATION EVOLVED
To support the remarkable pace and scale of these research fields, RNA extraction protocols need to be efficient and reliable. One method – acid-guanidinium-phenol-chloroform extraction – has become the “gold standard” for high-purity RNA extraction.
The original protocol for acid-guanidium-phenol-chloroform extraction was published in 1987.4 This single-step protocol, which eliminated a more time-consuming method and low-throughput ultracentrifugation step, was revolutionary.5 Overall it reduced the time for purification into a 4 hour process, providing “…both high yield and purity of undegraded RNA preparations.”4,5
At the beginning of purification, the acidic solution is added to lyse the sample, then centrifuged, separating the resulting mixture into three distinct phases: aqueous, interphase, and organic, from top-to-bottom. The RNA stays in the upper aqueous phase, while DNA and proteins remain in the lower two phases, along with cellular debris. This protocol has remained largely unchanged for the past few decades and the key isolation reagent, a monophasic solution of phenol and guanidine isothiocyanate, is commercially available under the brand names TRIzol® or Tri-Reagent®.6 These reagents both lyse a wide range of cell types, releasing RNA for extraction, and prevents RNA degradation by inhibiting (most) RNases. It also enables two other protocols (hence the “tri-“ prefix in the brand name) to be run in parallel or subsequently using the interphase and solid organic phase for genomic DNA and protein purification, respectively.7
RNA EXTRACTION FROM TRIZOL®
While RNA extraction with TRIzol® can seem relatively straightforward, there are a few key challenges that many first-time extractors can face.
Set Aside the Right Amount of Time
While current RNA extraction protocols don’t require 4 hours (as stated in the original 1987 paper), you will need about an hour, from sample lysis to resuspension of the dried RNA pellet. All the steps in the protocol are done manually, so the total extraction time largely depends on how many samples you have to process.
One of the major bottlenecks can be the centrifugation steps. Most microcentrifuges can hold between 20 to 30 samples, so if you’re processing more samples than that, you may have to run your RNA extraction in batches, which can significantly increase your extraction time.
Get Your Reagents Prepared & Know the Steps
There are four major steps in TRIzol® RNA extraction and a handful of reagents you’ll need for each:
- Cell lysis and phase separation: TRIzol® (or an acid-guanidinium-phenol based reagent) is required to lyse your sample, and chloroform is used for phase separation (as described above) of RNA (aqueous phase) from DNA (inter-/organic phase).
- RNA precipitation: The aqueous phase is transferred to a new tube, and isopropanol is added to precipitate RNA.
- RNA washing: Precipitated RNA is pelleted through centrifugation and then washed with 70 – 75% ethanol.
- RNA solubilization: Pellet resuspension requires either water or a buffered solution like TE. There are other solutions that you can use to resuspend your RNA, depending on your downstream application. Be sure to store your RNA at -70°C if you plan to use it later.
Tips & Tricks
As you prepare your reagents, one key consideration is keeping them RNase-free. Despite what is published in the original 1987 paper, some RNases, like those in the RNase A family, are stabilized by disulfide bonds and remain active in TRIzol®.8 If you were to speak to any RNA extraction pro, they would tell you that these can wreak havoc on your purification and leave you with low quality, degraded RNA.
To avoid this, try using stabilization reagents or prepare a special RNase-free zone, where all reagents, tubes, pipette tips, and pipettes are RNase-free. In addition, your skin can be one of the sources of RNase contamination during an extraction, so try to minimize contact before, during, and after your RNA extraction.9
Beware of Phase Separation
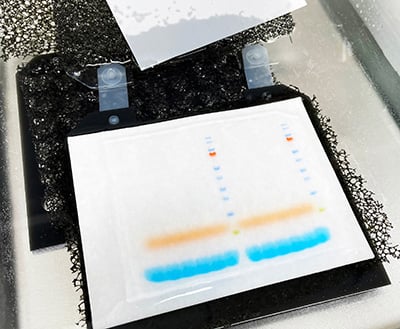
The major “make-or-break” step during RNA extraction is phase separation (Figure 1). It requires dexterity and careful pipetting to remove as much of the aqueous (RNA) phase as possible, without disturbing the DNA-rich interphase.
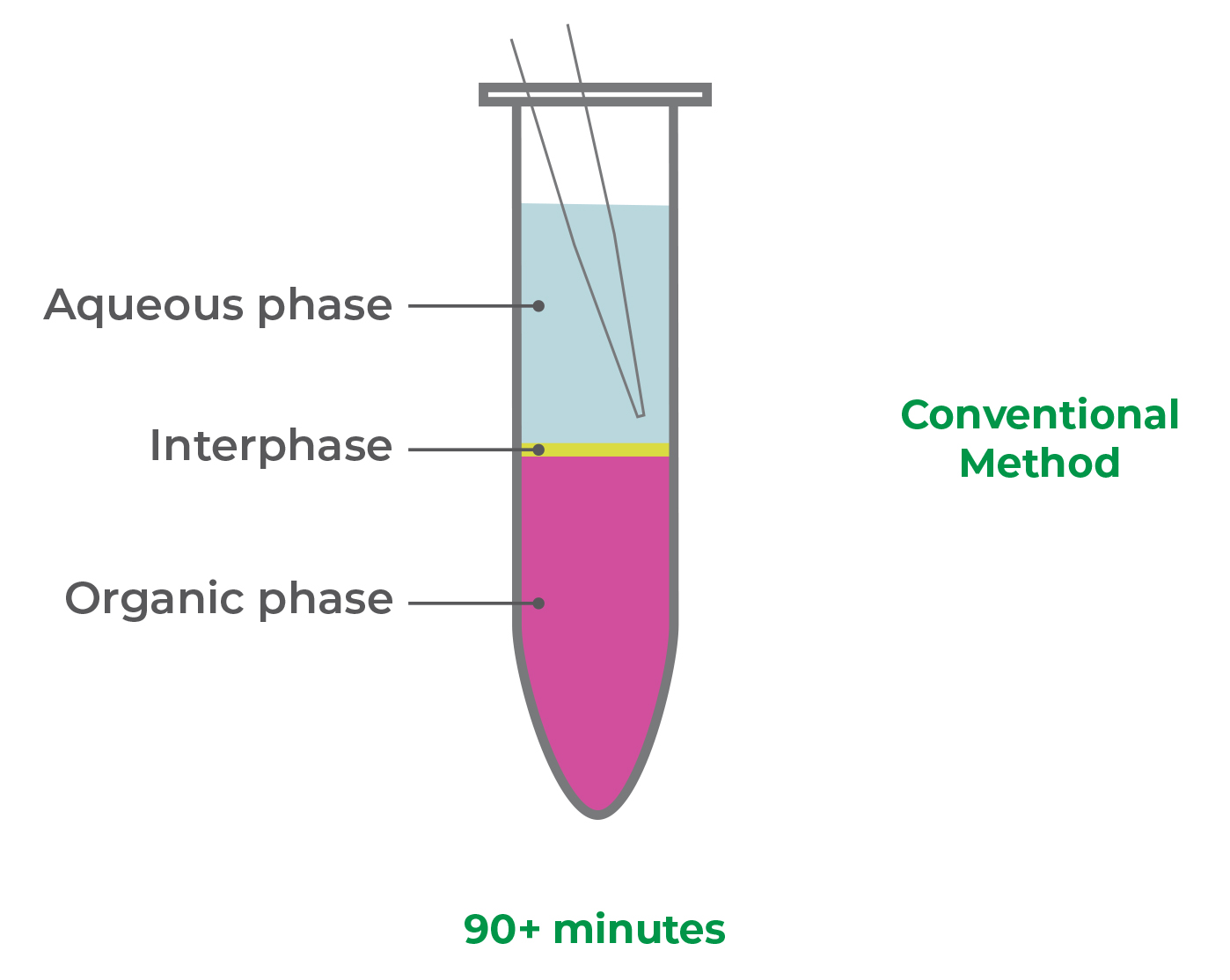 Figure 1. Removal of the aqueous (RNA) phase, without disturbing the inter- and/or organic phases, is a challenging, “make-or-break” step in RNA extraction from TRIzol.
Figure 1. Removal of the aqueous (RNA) phase, without disturbing the inter- and/or organic phases, is a challenging, “make-or-break” step in RNA extraction from TRIzol.
By disturbing the interphase, your purified RNA can be contaminated with genomic DNA, protein, and/or phenol, which can complicate quantification, reduce RNA quality, and inhibit downstream applications. This makes the phase separation step a major source of inconsistencies such as impurities and yield variability across samples, resulting in inaccurate data analysis and low reproducibility.
Furthermore, if you determine that you have genomic DNA contamination, you may have to follow up your RNA extraction with a DNase I digestion, which can add additional time to your overall workflow.
Low Inputs & Small RNAs
RNA extraction from TRIzol® relies on precipitation of RNA using isopropanol. With small amounts of RNA, precipitation is less efficient and can result in extremely low yields or no RNA pellet at all. Be sure to follow the manufacturer’s instructions and add the recommended input number of cells or tissue weight.
Precipitation is also inefficient for extracting small RNAs, like microRNAs. There can be biased results in the amount of small RNAs recovered.10 RNA extraction by phase separation was shown to selectively enrich for some species of small/miRNAs, leading to bias in downstream analysis. You can try using carriers, such as glycogen, to promote precipitation and improve your yield.11 Alternately, there are commercially available RNA purification kits specifically designed for small RNAs.
THE BEST TIP: FIND AN EASIER WAY TO EXTRACT RNA FROM TRIZOL®
While TRIzol® extraction is still considered the “gold standard,” there are a lot of limitations that make it unsuitable for your experiments.
Zymo Research’s Direct-zol RNA Kits offer you a powerful alternative, reducing the entire RNA purification protocol from an hour to just seven minutes.
Starting with a sample resuspended in TRIzol®, there are just three simple steps: bind, wash, and elute RNA.
First, add an equal volume of ethanol (1:1) to the lysed sample in TRIzol® and mix. Bind RNA to the silica-based column, wash the column, then elute RNA (Figure 2).
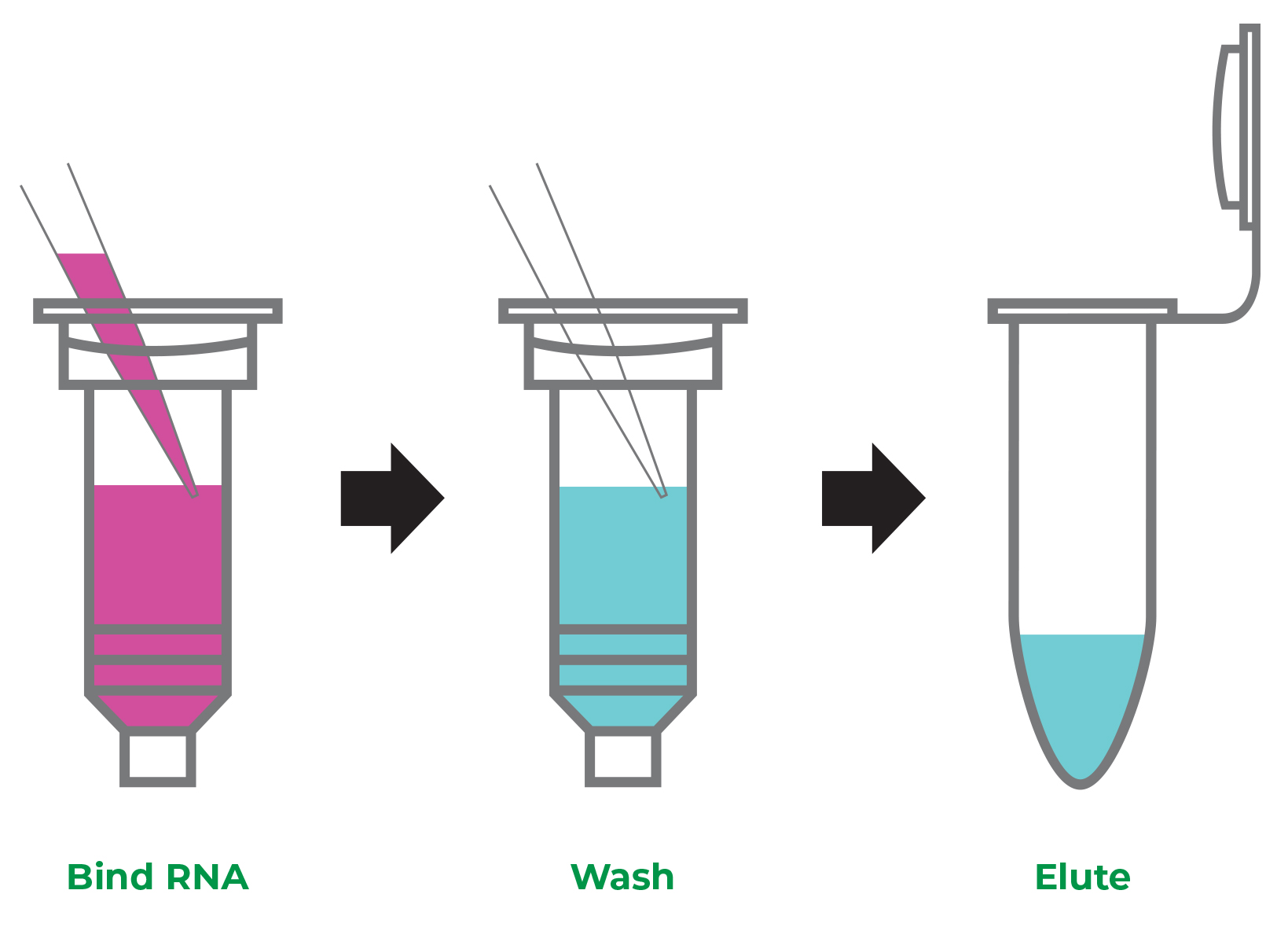 Figure 2. No more precision pipetting. With Direct-zol, just bind, wash, and elute the RNA.
Figure 2. No more precision pipetting. With Direct-zol, just bind, wash, and elute the RNA.
The Direct-zol RNA kit includes DNase I, so you can also rapidly remove contaminating DNA. It eliminates the need for chloroform (no phase separation) and concerns over phenol contamination or bias in small RNA recovery (Figure 3).
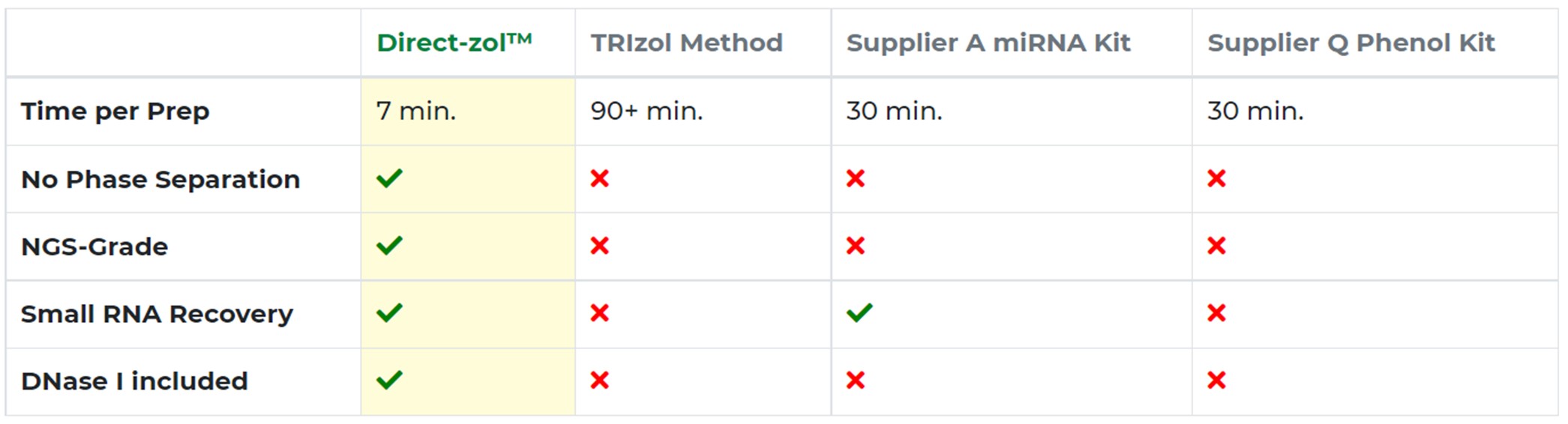 Figure 3. Direct-zol delivers higher-quality RNA preps in a fraction of the time of other methods or kits.
Figure 3. Direct-zol delivers higher-quality RNA preps in a fraction of the time of other methods or kits.
At the end of the Direct-zol extraction process, the result is high-purity, high yield RNA – all the RNA. Direct-zol captures up to 4-times smaller/microRNAs than the traditional method and can deliver even picograms of RNA (Figure 4).
Three steps, seven minutes, and done.
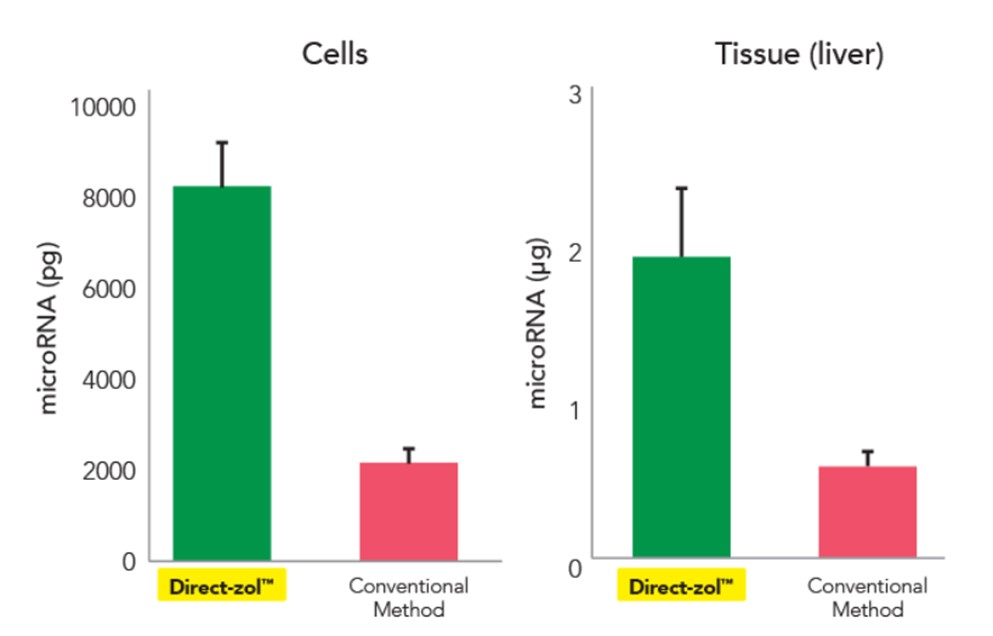 Figure 4. Direct-zol allows for 4x the recovery of microRNAs.
Figure 4. Direct-zol allows for 4x the recovery of microRNAs.
What is even more exciting is the possibility for high-throughput automation. Direct-zol has a bead-based solution which can be adapted for any automated platform including Tecan, Hamilton, and Thermo’s KingFisher®. If you are interested, Zymo Research provides free automation scripts with technical support, as well as column-based sample kits for evaluation.
The future of diagnostics, drug discovery, vaccine development and more begins with high-quality RNA isolation, and Zymo Research has the tools to help your lab deliver the next big biomedical breakthrough.